Software
arg-lmm
arg-lmm provides efficient computational tools for complex trait analysis using Ancestral Recombination Graphs (ARGs).
Quickdraws
Quickdraws is a scalable method to perform genome-wide association studies (GWAS) for quantiative and binary traits.
[software] [manual] [RAP tutorial]
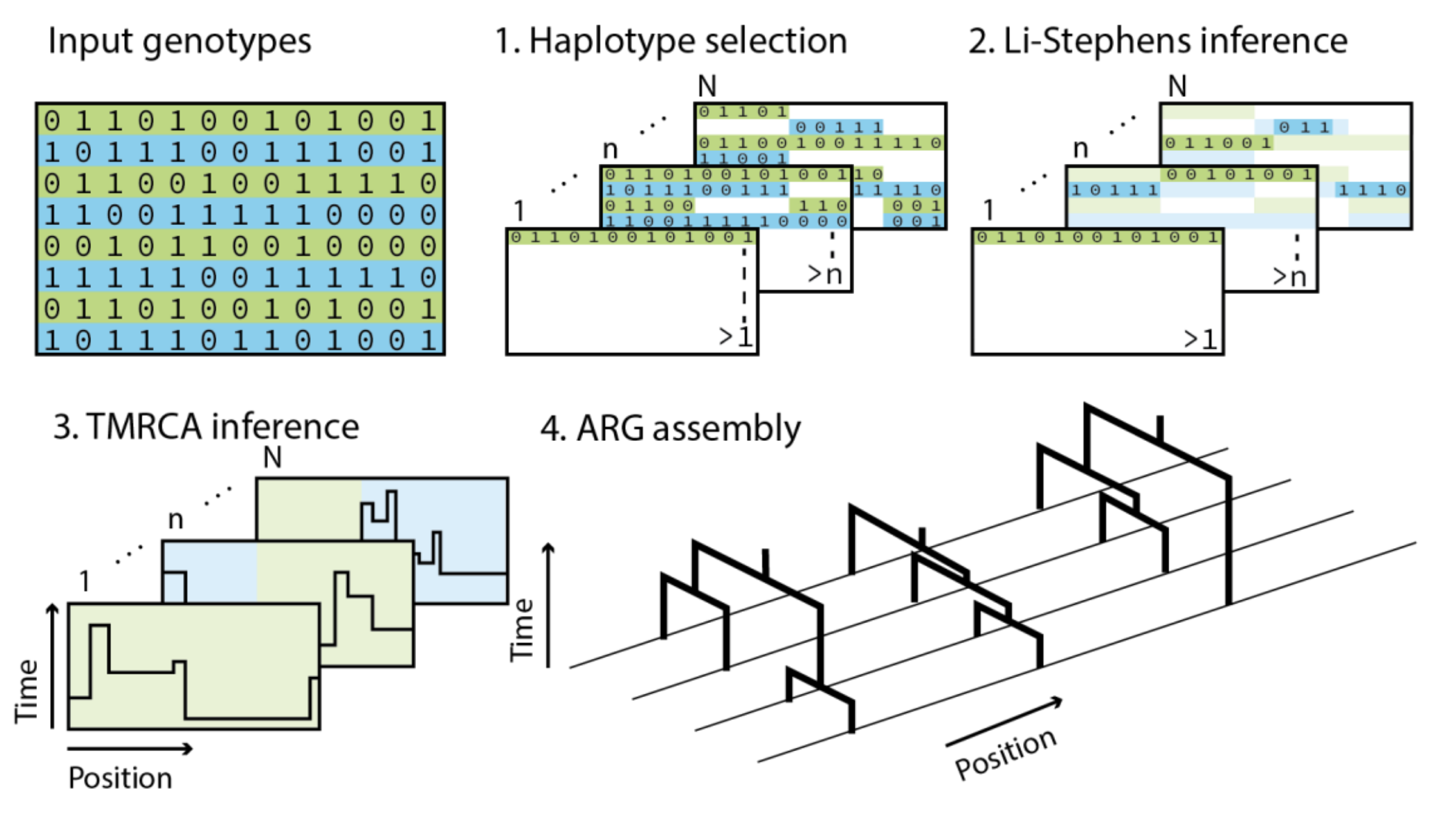
Threads
Threads is a threading-based method to reconstruct the ancestral recombination graph (ARG) from large datasets of sequenced, imputed, or genotyped genomes and to perform ARG-based genotype imputation.
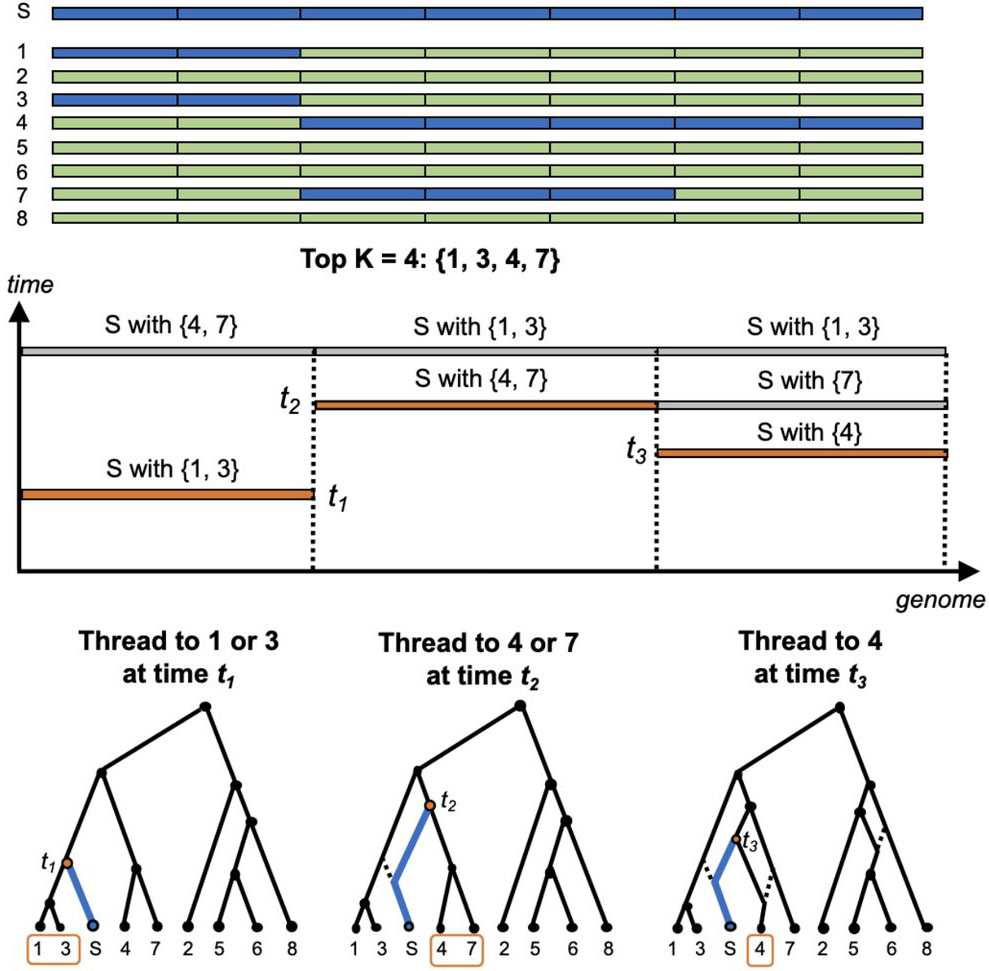
ARG-Needle
ARG-Needle is an algorithm to reconstruct the genealogical history (ancestral recombination graph, ARG) of large collections of sequenced or genotyped genomes. ARG-Needle-lib is a library that can be used to perform genealogy-wide association and other analyses. This package also implements the ASMC-clust ARG reconstruction algorithm described in the manuscript.
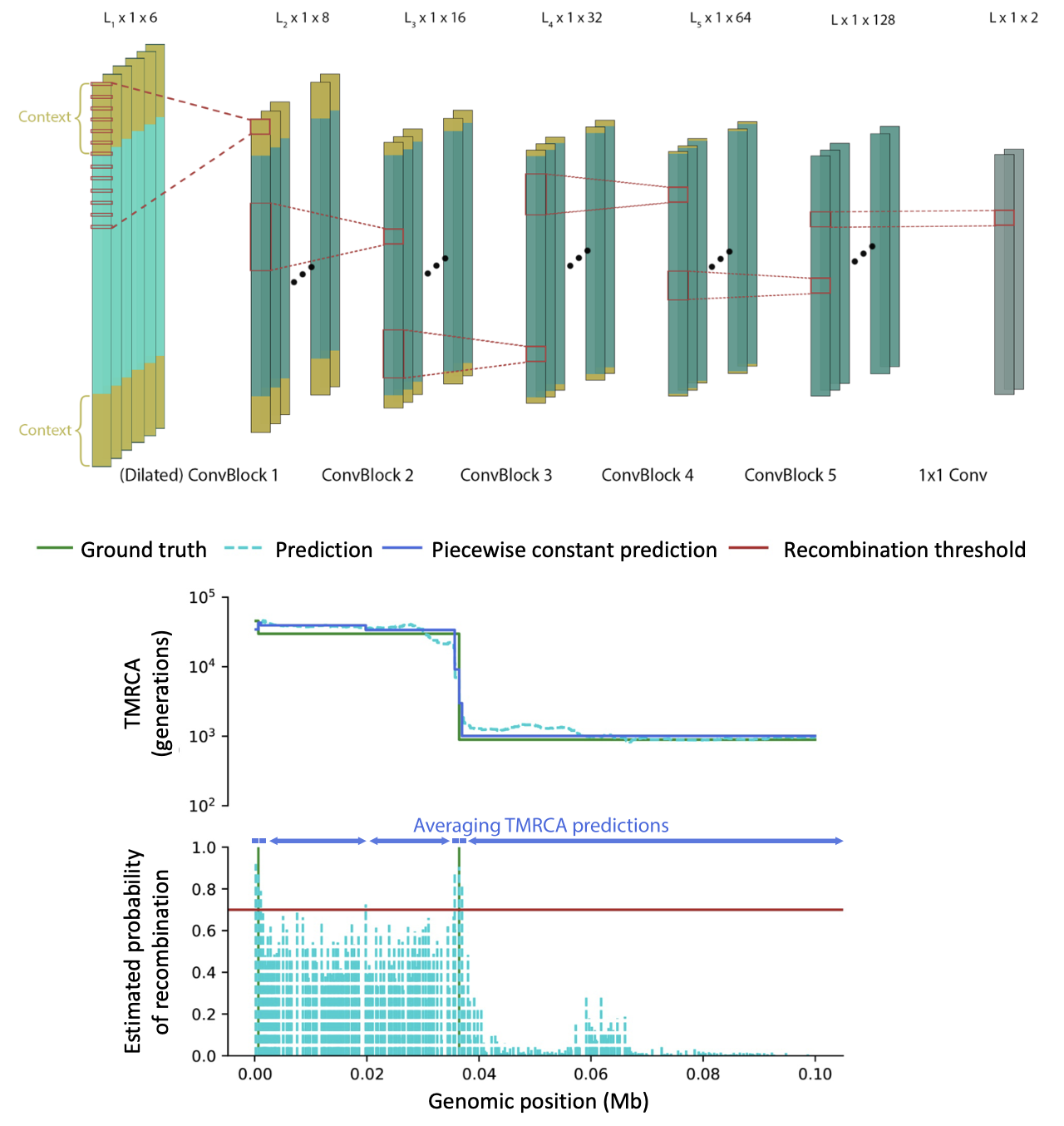
CoalNN
CoalNN is a likelihood-free method, based on convolutional neural networks, for the estimation of TMRCAs and allele ages in sequencing, array, and imputed genomic data.
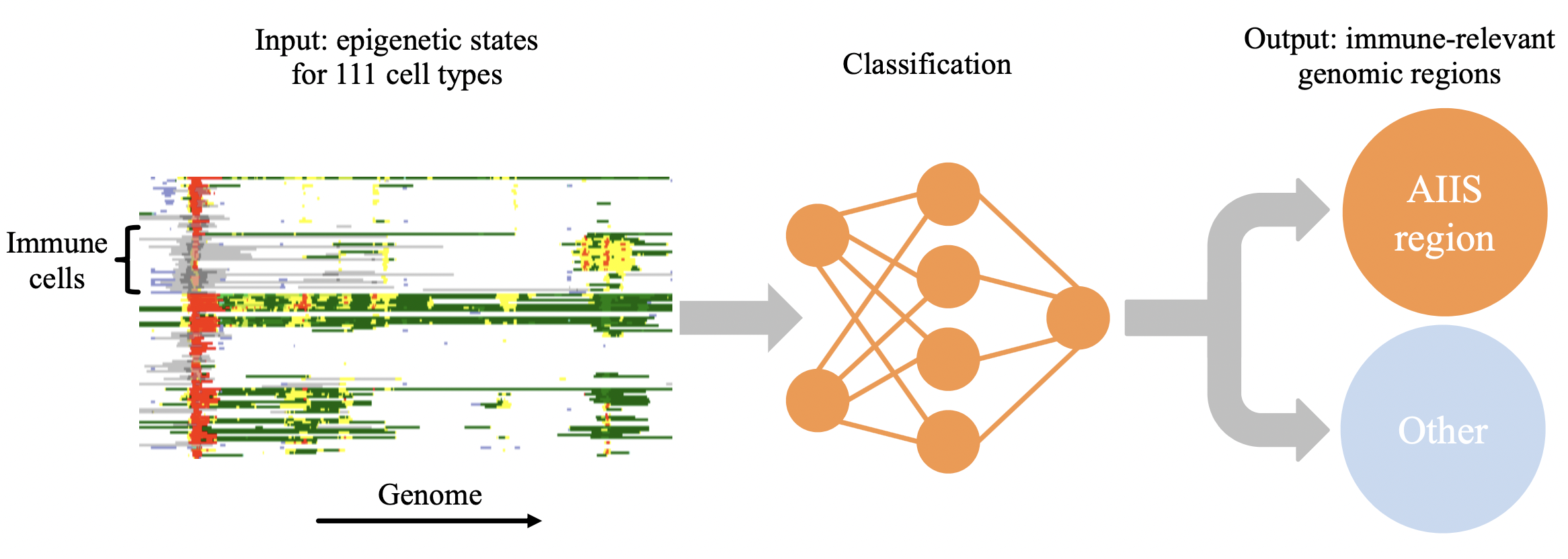
EpiNN
EpiNN uses a classification algorithm (convolutional neural network) trained with publicly available epigenetic data to recognize genomic regions likely to play a role in the adaptive and innate immune system.
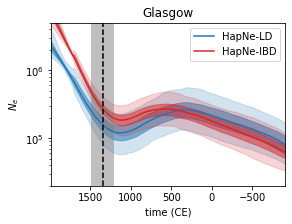
HapNe
HapNe is a method to reconstruct recent past demographic fluctuations in modern and ancient data.
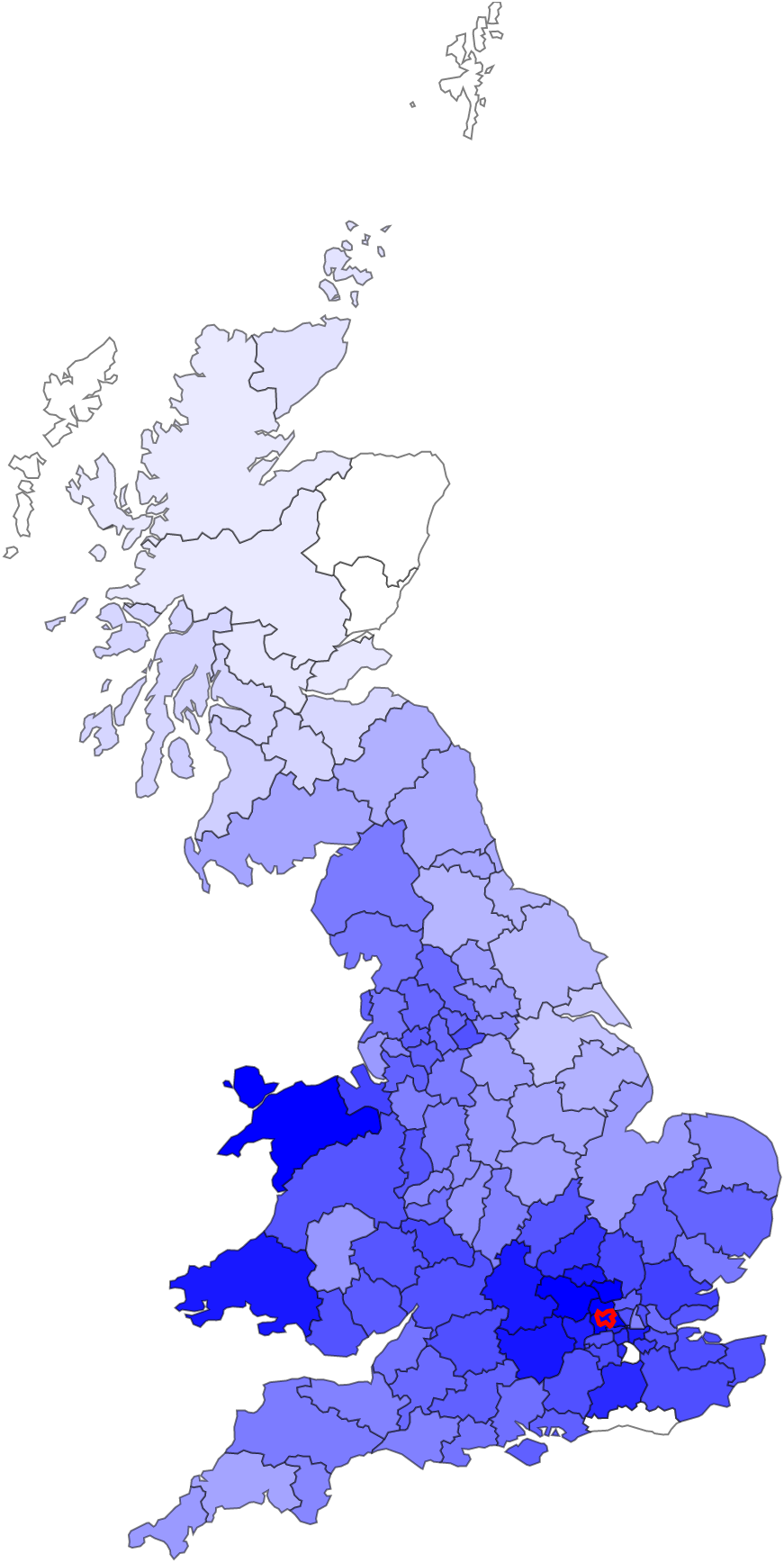
FastSMC
FastSMC is a method to quickly and accurately estimate pairwise identical-by-descent (IBD) regions in the genome. FastSMC estimates the age of IBD segments and scales to large biobank datasets.
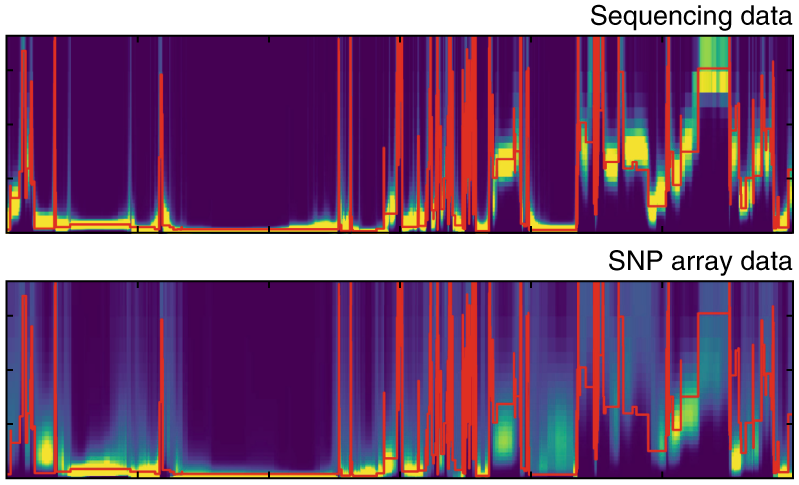
ASMC
The Ascertained Sequentially Markovian Coalescent is a method to efficiently estimate pairwise coalescence times along the genome. It can be run using SNP array or whole-genome sequencing (WGS) data.
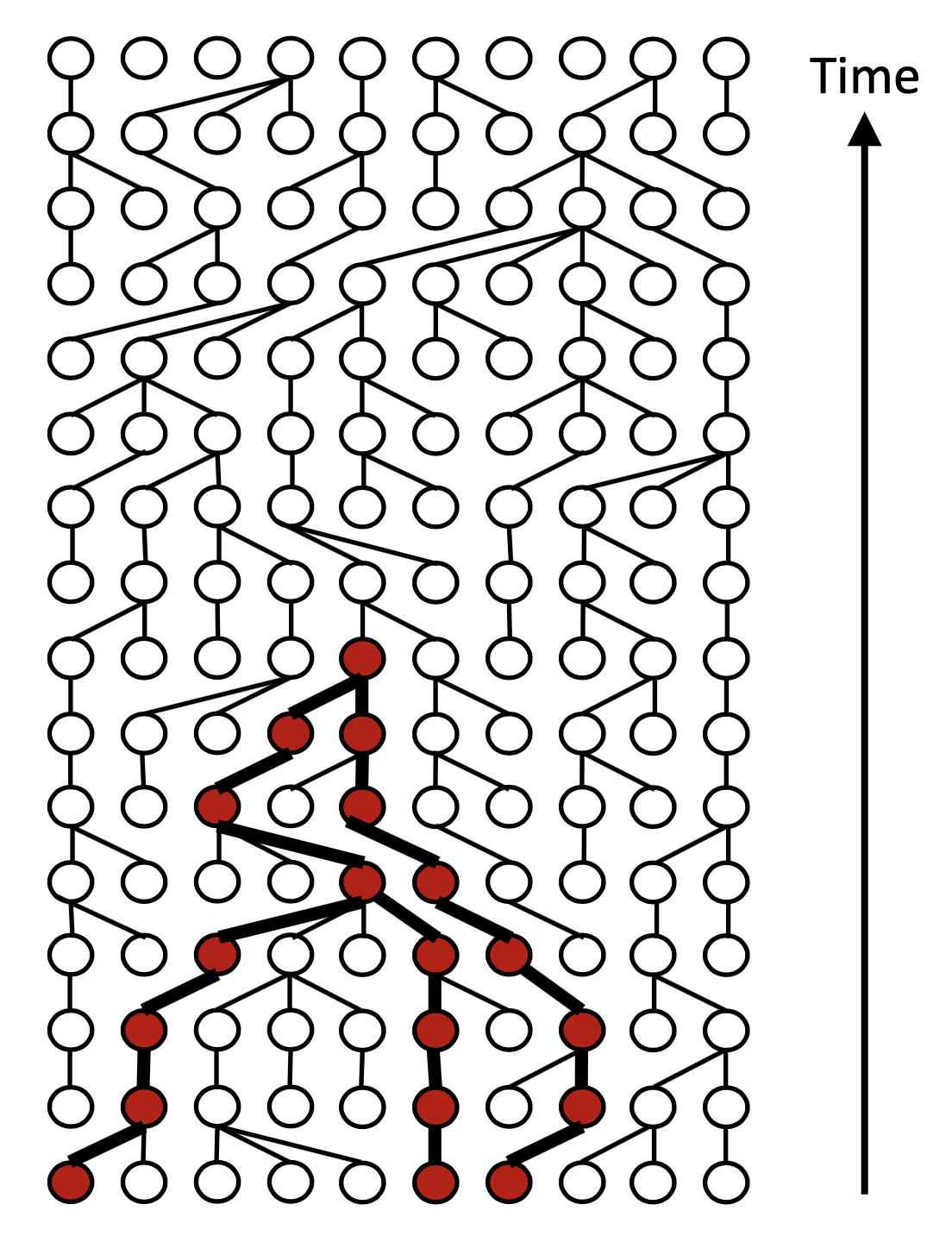
ARGON
ARGON is a fast simulator of genetic data that samples from the discrete time Wright-Fisher process (DTWF) backwards in time. It first builds an ancestral recombination graph (ARG) and then uses it to generate genetic data or to efficiently compute pairwise haplotype sharing (IBD) statistics.
IBDmut
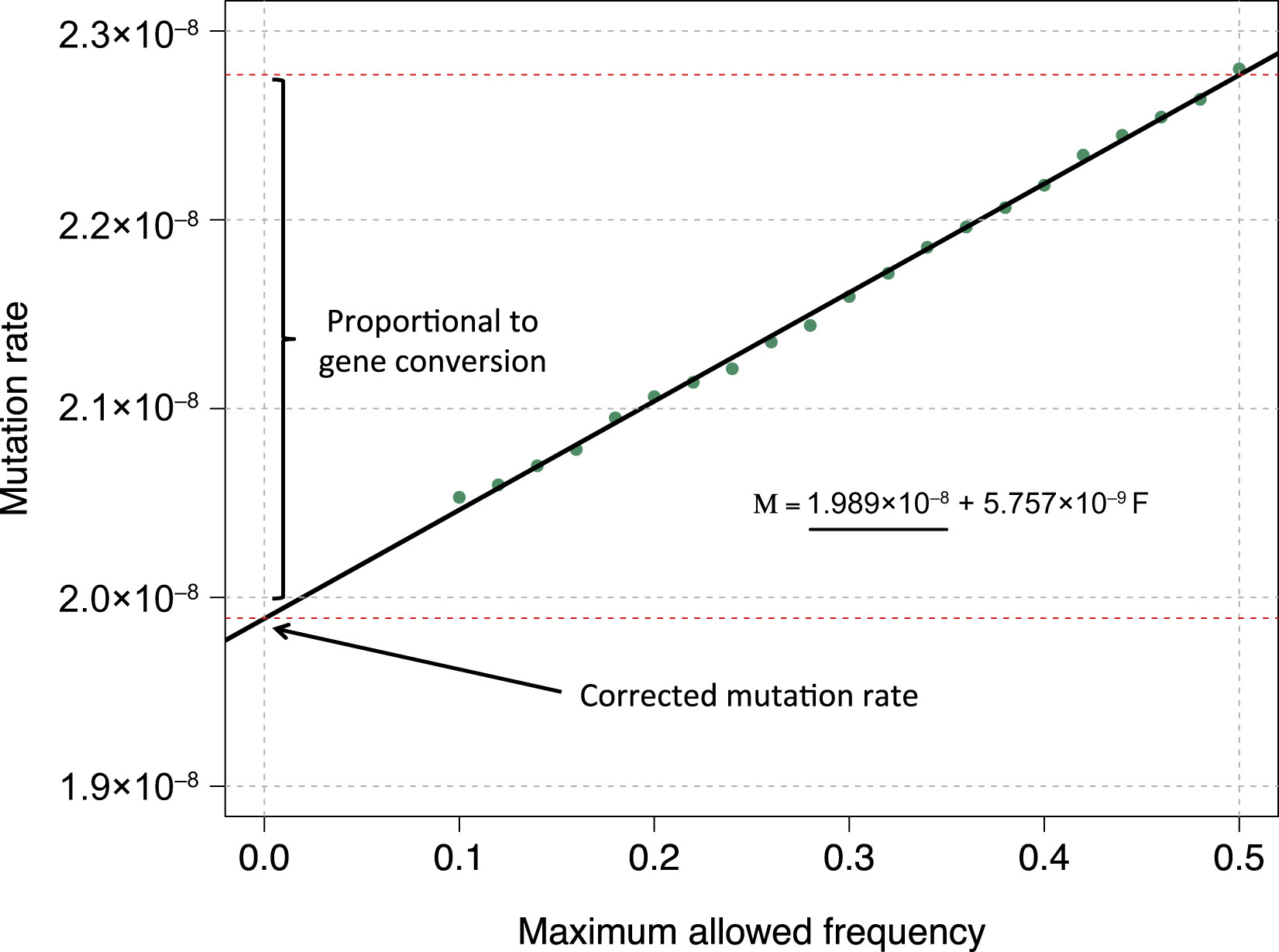
IBDmut estimates mutation and non-crossover gene conversion rates using identical-by-descent (IBD) segments shared by distant relatives within a population.